Identifying COMMON links of fMRI data
Thank you for using this package for identifying COMMON links --[download here]
.Fig. 1. An example of a network of fMRI data identified by COMMON [1].
EASY TO RUN
This is a Matlab GUI for identifying common links in functional networks from fMRI data. It is still under testing, so any feedback from you would be more than welcome. Free for acadamic use only. In the current working path in MatLab, the GUI is avaliable if you type in
COMMON
Then the use of this toolbox should be straightforward.
METHODS
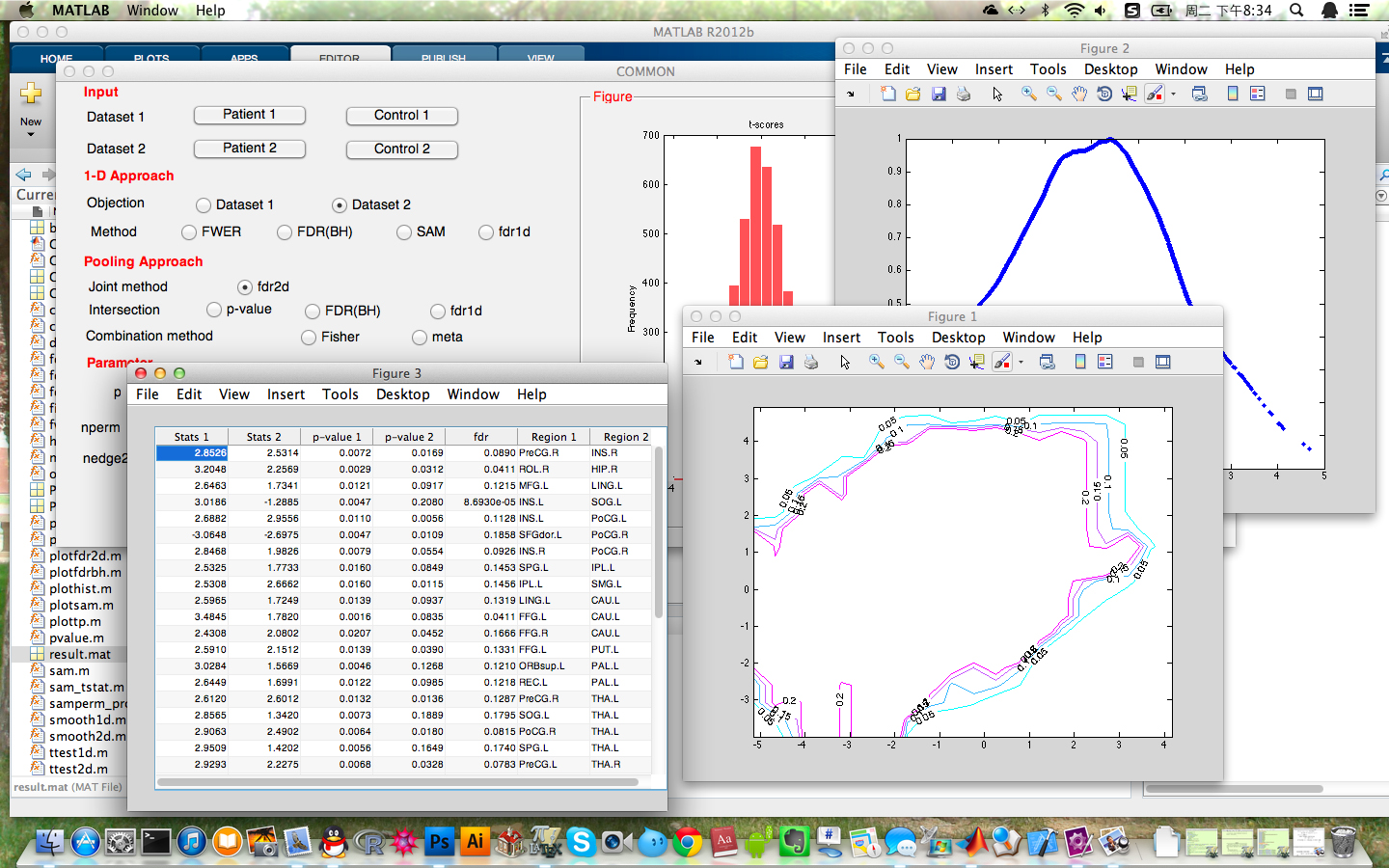
The interface looks like the following, including some popular methods on 1-D and 2-D fdr, like FDR(B-H), 1-d local false discovery rate and SAM.
The procedure using to identify the "Common" links is as follows,
1) With the help of 2-D fdr approach, taking 0.1 as threshold, find out the significant subset denoted by A.
2) Implementing 1-D method (1-D fdr, FDR (B-H), SAM et. al) on the two datasets (attributions) respectively with a threshold α, obtain the significant subset B from the first dataset (attribution) and C from the second dataset (attribution).
3) Find out the common links A cap B cap C.
The results are shown in tables and figures. The results are also saved in 'result.mat' in the current working path. All the returns of programs in the package is a struct, named 'output', including following details, output, a struct, .fdr, the local fdr for the links .stat, the statistics for the links .pvalue, the p-values for the links .p0, the prior proportion of the null links used in the estimation .th, the matrix for result with fdr < q.
Fig. 2. A screenshot of the toolbox.
CONTACT
This package is developed by Jianfeng Feng group, in accompany of papers
[1] A Statistical Approach for Trans-attibution Model submitted, 2013.
-----------------------------------------------------------
e-mail: Email: xjgansdu@163.com, last modified by XJ Gan at 01:10 12/23/2013.